Welcome to the documentation for Escher¶
Escher is a web-based tool for building, viewing, and sharing visualizations of metabolic pathways. These ‘pathway maps’ are a great way to contextualize data about metabolism. The easiest way to use Escher is to browse or build maps on the Escher website.
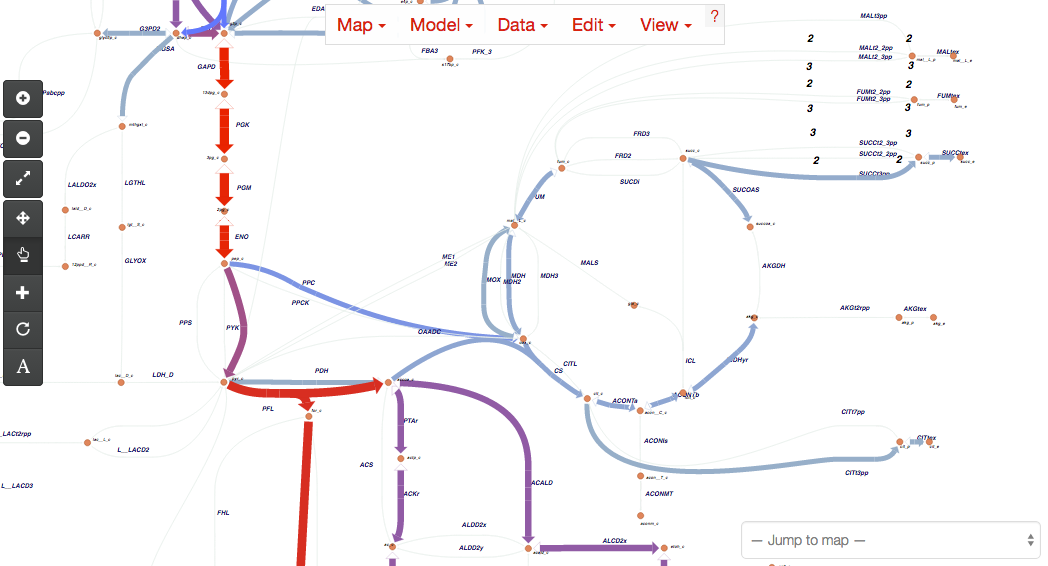
Features¶
- View pathway maps in any modern web browser
- Build maps using the content of genome-scale metabolic models
- Visualize data on reactions, genes, and metabolites
- Full text search
- Detailed options for changing colors, sizes, and more, all from the web browser
- View maps inside the IPython Notebook
- Embed maps within any website, with minimal dependencies (escher.js, d3.js, and optionally Twitter Bootstrap)
Supported browsers¶
We recommend using Google Chrome for optimal performance, but Escher will also run in the latest versions of Firefox, Internet Explorer, and Safari (including mobile Safari).
Installation¶
Escher can be used without any installation by visiting the Escher website. However, you can install escher if you would like to (1) run Escher offline, (2) include your own maps and models in the launch page, (3) view Escher maps in an IPython Notebook, or (4) modify the source code.
To install the latest stable version of Escher, run:
pip install escher
For more information, see the documentation on Escher in the IPython Notebook and Developing with Escher.
Contents¶
License¶
Escher is licensed under the MIT license.